Predicting Cell Phase
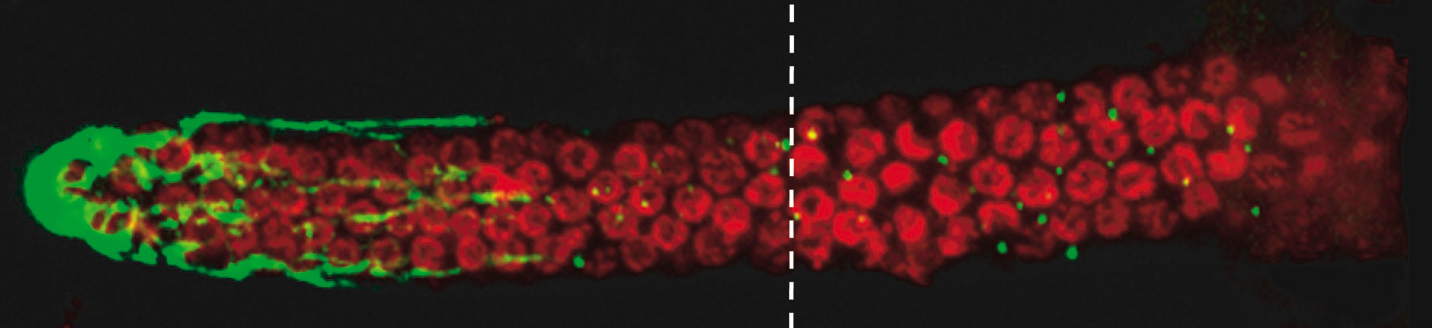
Fig. 1 The end of a C. elegans worm as seen by confocal microscope. The stem cells have been stained red. Different reagents bind and highlight the DNA shape can which can be used to predict the cell phase. The large green blob on the left is the distal tip cell, which generates the stem cells that flow down the worm differentiating into their various roles.
Add computer vision
Stem cells are interesting because they can divide and specialize. Control of stem cell development holds the promise of engineering new organs. While the use of human stem cells in research is fraught with ethical complications, `C. elegans`_ worms are small, translucent and have a short life span making them ideal models for studying stem cells. Confocal microscopes and DNA staining let us create 3D images of their DNA similar to that in Fig. 1.
An animal cell cycles through four stages of development: growth (G1), synthesize new DNA (S), more growth (G2), and splitting or mitosis (M). Staining applies a reagent doped with a fluorescent molecule that binds to a desired target. It is possible to determine which cells are in a certain phase by staining them with the corresponding reagent; unfortunately, using multiple stains causes complications.
The goal of this project is to use the DNA shape to classify the cell's phase.
The procedure:
Stain cell DNA and one phase.
Image worms using a confocal microscope.
Segment the image into individual cells.
Run a variety of image processing filters on the microscopy image to create a feature vector.
Partition data into training, validation, and test sets.
Train a support vector machine on the training set.
Optimize parameters on the validation set.
Test its accuracy on the test set.
The code I inherited was a case study in poor programming practices:
No documentation
Inconsistent indentation
Significant code redundancy
Fragile: changing the database selection criteria required modifying nearly every file in the project.
Matlab was a poor choice of languages for an image database and supervised learning algorithm.
Database key uniqueness issues
During my limited time on the project, I made the following improvements:
Created a git repository
Improved the directory structure: source code, data, documentation
Refactored the code to remove redundancy
Reindexed the database to accomodate project growth
Wrote a documenttion for future users
Improved prediction accuracy from 85% to 90%